From Multilayer Networks to Deep Graphs¶
[ipython notebook] [python script]
In this tutorial we exemplify the representation of multilayer networks (MLNs) by deep graphs and demonstrate some of the advantages of deepgraph’s network representation.
We start by converting the Noordin Top Terrorist MLN into a graph g - comprised of two DataFrames, a node table g.v and an edge table g.e - that corresponds to the supra-graph representation of the multilayer network.
We then partition the graph g by the information attributed to its layers, resulting in different supergraphs on the partition lattice of g that correpsond to different representations of a MLN (including its tensor representation).
In the next part, we demonstrate how additional information that might be at hand or computed during the analysis can be used to induce further supergraphs, or metaphorically speaking, how additional information corresponds to “hidden layers” of a MLN.
Finally, we briefly show how to use the nodes’ properties to partition the edges of a MLN.
** References **
For a short summary of the multilayer network representation, see Appendix C of the Deep Graphs paper.
For a more in-depth introduction to MLNs, I recommend the following papers:
- Multilayer Networks (review paper of MLNs)
- The Structure and Dynamics of Multilayer Networks (review paper of MLNs)
- Mathematical Formulation of Multilayer Networks (tensor formalism for MLNs)
For a discussion of how Deep Graphs relates to the multilayer network representation, see Sec. IV B and Appendix D of the Deep Graphs paper.
The Noordin Top Terrorist Data¶
[high-res version] [python plot script]
The data we use in this tutorial is the Noordin Top Terrorist Network, which has previously been represented as a multilayer network (e.g., http://arxiv.org/abs/1308.3182)
It includes relational data on 79 Indonesian terrorists belonging to the so-called Noordin Top Terrorist Network.
For information about the individual’s attributes and their relations, see http://www.thearda.com/archive/files/codebooks/origCB/Noordin%20Subset%20Codebook.pdf and http://arxiv.org/pdf/1308.3182v3.pdf.
Preprocessing¶
We download the data from here, and process it into two pandas DataFrames, a node table and an edge table. The preprocessing is quite lengthy, so you might want to proceed directly to the next section.
First of all, we need to import some packages
# data i/o
import os
import subprocess
import zipfile
# for plots
import matplotlib.pyplot as plt
# the usual
import numpy as np
import pandas as pd
import deepgraph as dg
# notebook display
%matplotlib inline
pd.options.display.max_rows = 10
pd.set_option('expand_frame_repr', False)
Preprocessing the Nodes¶
# zip file containing node attributes
os.makedirs("tmp", exist_ok=True)
get_nodes_zip = ("wget -O tmp/terrorist_nodes.zip "
"https://sites.google.com/site/sfeverton18/"
"research/appendix-1/Noordin%20Subset%20%28ORA%29.zip?"
"attredirects=0&d=1")
subprocess.call(get_nodes_zip.split())
# unzip
zf = zipfile.ZipFile('tmp/terrorist_nodes.zip')
zf.extract('Attributes.csv', path='tmp/')
zf.close()
# create node table
v = pd.read_csv('tmp/Attributes.csv')
v.rename(columns={'Unnamed: 0': 'Name'}, inplace=True)
# create a copy of all nodes for each layer (i.e., create "node-layers")
# there are 10 layers and 79 nodes on each layer
v = pd.concat(10*[v])
# add "aspect" as column to v
layer_names = ['Business', 'Communication', 'O Logistics', 'O Meetings',
'O Operations', 'O Training', 'T Classmates', 'T Friendship',
'T Kinship', 'T Soulmates']
layers = [[name]*79 for name in layer_names]
layers = [item for sublist in layers for item in sublist]
v['layer'] = layers
# set unique node index
v.reset_index(inplace=True)
v.rename(columns={'index': 'V_N'}, inplace=True)
# swap columns
cols = list(v)
cols[1], cols[10] = cols[10], cols[1]
v = v[cols]
# get rid of the attribute columns for demonstrational purposes,
# will be inserted again later
v, vinfo = v.iloc[:, :2], v.iloc[:, 2:]
Preprocessing the Edges¶
# paj file containing edges for different layers
get_paj = ("wget -O tmp/terrorists.paj "
"https://sites.google.com/site/sfeverton18/"
"research/appendix-1/Noordin%20Subset%20%28Pajek%29.paj?"
"attredirects=0&d=1")
subprocess.call(get_paj.split())
# get data blocks from paj file
with open('tmp/terrorists.paj') as txtfile:
comments = []
data = []
part = []
for line in txtfile:
if line.startswith('*'):
# comment lines
comment = line
comments.append(comment)
if part:
data.append(part)
part = []
else:
# vertices
if comment.startswith('*Vertices') and len(line.split()) > 1:
sublist = line.split('"')
sublist = sublist[:2] + sublist[-1].split()
part.append(sublist)
# edges or partitions
elif not line.isspace():
part.append(line.split())
# append last block
data.append(part)
# extract edge tables from data blocks
ecomments = []
eparts = []
for i, c in enumerate(comments):
if c.startswith('*Network'):
del data[0]
elif c.startswith('*Partition'):
del data[0]
elif c.startswith('*Vector'):
del data[0]
elif c.startswith('*Arcs') or c.startswith('*Edges'):
ecomments.append(c)
eparts.append(data.pop(0))
# layer data parts (indices found manually via comments)
inds = [11, 10, 5, 6, 7, 8, 0, 1, 2, 3]
eparts = [eparts[ind] for ind in inds]
# convert to DataFrames
layer_frames = []
for name, epart in zip(layer_names, eparts):
frame = pd.DataFrame(epart, dtype=np.int16)
# get rid of self-loops, bidirectional edges
frame = frame[frame[0] < frame[1]]
# rename columns
frame.rename(columns={0: 's', 1: 't', 2: name}, inplace=True)
frame['s'] -= 1
frame['t'] -= 1
layer_frames.append(frame)
# set indices
for i, e in enumerate(layer_frames):
e['s'] += i*79
e['t'] += i*79
e.set_index(['s', 't'], inplace=True)
# concat the layers
e = pd.concat(layer_frames)
# edge table as described in the paper
e_paper = e.copy()
# alternative representation of e
e['type'] = 0
e['weight'] = 0
for layer in layer_names:
where = e[layer].notnull()
e.loc[where, 'type'] = layer
e.loc[where, 'weight'] = e.loc[where, layer]
e = e[['type', 'weight']]
DeepGraph’s Supra-Graph Representation of a MLN, \(G = (V, E)\)¶
Above, we have processed the downloaded data into a node table v and
an edge table e, that correspond to the supra-graph representation
of a multilayer network. This is the preferred representation of a MLN
by a deep graph, since all other representations are entailed in the
supra-graph’s partition lattice, as we will demonstrate below.
g = dg.DeepGraph(v, e)
print(g)
<DeepGraph object, with n=790 node(s) and m=1014 edge(s) at 0x7fb8e13499e8>
Let’s have a look at the node table first
print(g.v)
V_N layer
0 0 Business
1 1 Business
2 2 Business
3 3 Business
4 4 Business
.. ... ...
785 74 T Soulmates
786 75 T Soulmates
787 76 T Soulmates
788 77 T Soulmates
789 78 T Soulmates
[790 rows x 2 columns]
As you can see, there are 790 nodes in total. Each of the 10 layers,
print(g.v.layer.unique())
['Business' 'Communication' 'O Logistics' 'O Meetings' 'O Operations'
'O Training' 'T Classmates' 'T Friendship' 'T Kinship' 'T Soulmates']
is comprised of 79 nodes. Every node has a feature of type V_N,
indicating the individual the node belongs to, and a feature of type
layer, corresponding to the layer the node belongs to. Each of the
790 nodes corresponds to a node-layer of the MLN representation of this
data.
The edge table,
print(g.e)
type weight
s t
9 67 Business 2.0
69 Business 1.0
77 Business 1.0
11 61 Business 1.0
20 59 Business 1.0
... ... ...
733 769 T Soulmates 1.0
755 769 T Soulmates 1.0
787 T Soulmates 1.0
771 788 T Soulmates 1.0
783 788 T Soulmates 1.0
[1014 rows x 2 columns]
is comprised of 1014 edges between the nodes in v. Each edge has two relations. The first relation (of type type) is determined by the tuple of features \((layer_i, layer_j)\) of the adjacent nodes \(V_i\) and \(V_j\). The second relation (of type weight) indicates the “weight” of the connection.
This representation of the edges of a MLN deviates from the one you can find in the paper, which is described in the last section.
There are 10 types of relations in the above edge table
g.e['type'].unique()
array(['Business', 'Communication', 'O Logistics', 'O Meetings',
'O Operations', 'O Training', 'T Classmates', 'T Friendship',
'T Kinship', 'T Soulmates'], dtype=object)
which - in the case of this data set - correspond to the layers of the
nodes. This is due to the fact that there are no inter-layer connections
in the Noordin Top Terrorist Network (such as, e.g., an edge from layer
Business to layer Communication would be). The edges here are
all (undirected) intra-layer edges (e.g., Business \(\rightarrow\)
Business, Operations \(\rightarrow\) Operations).
To see how the edges are distributed among the different types, you can simply type
g.e['type'].value_counts()
O Operations 267
Communication 200
T Classmates 175
O Training 147
T Friendship 91
O Meetings 63
O Logistics 29
T Kinship 16
Business 15
T Soulmates 11
Name: type, dtype: int64
Let’s have a look at how many “actors” (nodes with at least one connection) there are within each layer
# append degree
gtg = g.return_gt_graph()
g.v['deg'] = gtg.degree_property_map('total').a
# how many "actors" are there per layer?
g.v[g.v.deg != 0].groupby('layer').size()
layer
Business 13
Communication 74
O Logistics 16
O Meetings 26
O Operations 39
O Training 38
T Classmates 39
T Friendship 61
T Kinship 24
T Soulmates 9
dtype: int64
For the purpose of this tutorial, the fact that the Noordin Top Terrorist Network is a MLN with only one aspect, and without inter-layer edges, is of little importance. The generalization of what we’re showing in the following to more general MLNs is straight-forward (and explained in detail in Appendix D of the paper).
Let’s illustrate the supra-graph representation of this MLN by a plot
# create graph_tool graph for layout
import graph_tool.draw as gtd
gtg = g.return_gt_graph()
gtg.set_directed(False)
# get sfdp layout postitions
pos = gtd.sfdp_layout(gtg, gamma=.5)
pos = pos.get_2d_array([0, 1])
g.v['x'] = pos[0]
g.v['y'] = pos[1]
# configure nodes
kwds_scatter = {'s': 1,
'c': 'k'}
# configure edges
kwds_quiver = {'headwidth': 1,
'alpha': .3,
'cmap': 'prism'}
# color by type
C = g.e.groupby('type').grouper.group_info[0]
# plot
fig, ax = plt.subplots(1, 2, figsize=(15, 7))
g.plot_2d('x', 'y', edges=True, C=C,
kwds_scatter=kwds_scatter,
kwds_quiver=kwds_quiver, ax=ax[0])
# turn axis off, set x/y-lim
ax[0].axis('off')
ax[0].set_xlim((g.v.x.min() - 1, g.v.x.max() + 1))
ax[0].set_ylim((g.v.y.min() - 1, g.v.y.max() + 1))
# plot adjacency matrix
adj = g.return_cs_graph().todense()
adj = adj + adj.T
inds = np.where(adj != 0)
ax[1].scatter(inds[0], inds[1], c='k', marker='.')
ax[1].grid()
ax[1].set_xlim(-1, 791)
ax[1].set_ylim(-1,791)
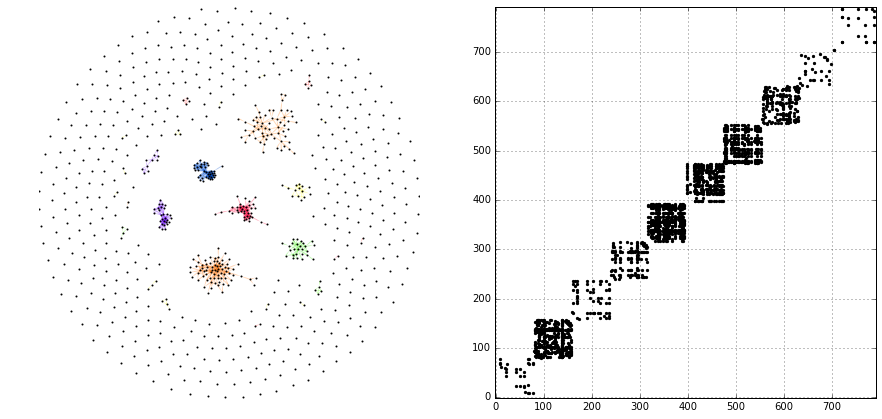
The supra-graph representation of a MLN is by itself a powerful representation and exploitable in various ways (see, e.g., section 2.3 of this paper). However, in the following, we will demonstrate how to use the additional information attributed to the layers of the MLN, in order to “structure” and partition the MLN into different representations.
Redistributing Information on the Partition Lattice of the MLN¶
Based on the types of features V_N and layer, we can now
redistribute the information contained in the supra-graph g. This
redistribution allows for several representations of the graph, which we
will demonstrate in the following.
The SuperGraph \(G^L = (V^L, E^L)\)¶
Partitioning by the type of feature layer leads to the supergraph
\(G^L = (V^L,E^L)\), where every supernode
\(V^{L}_{i^L} \in V^{L}\) corresponds to a distinct layer,
encompassing all its respective nodes. Superedges
\(E^{L}_{i^L, j^L} \in E^{L}\) with either \(i^L = j^L\) or
\(i^L \neq j^L\) correspond to collections of intra- and inter-layer
edges of the MLN, respectively.
# partition the graph
lv, le = g.partition_graph('layer',
relation_funcs={'weight': ['sum', 'mean', 'std']})
lg = dg.DeepGraph(lv, le)
print(lg)
<DeepGraph object, with n=10 node(s) and m=10 edge(s) at 0x7fb8e1349c50>
print(lg.v)
n_nodes
layer
Business 79
Communication 79
O Logistics 79
O Meetings 79
O Operations 79
O Training 79
T Classmates 79
T Friendship 79
T Kinship 79
T Soulmates 79
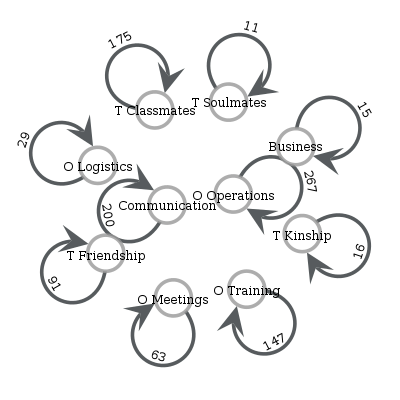
print(lg.e)
n_edges weight_sum weight_mean weight_std
layer_s layer_t
Business Business 15 16.0 1.066667 0.258199
Communication Communication 200 200.0 1.000000 0.000000
O Logistics O Logistics 29 58.0 2.000000 0.000000
O Meetings O Meetings 63 170.0 2.698413 1.612801
O Operations O Operations 267 574.0 2.149813 0.699107
O Training O Training 147 334.0 2.272109 0.763534
T Classmates T Classmates 175 175.0 1.000000 0.000000
T Friendship T Friendship 91 91.0 1.000000 0.000000
T Kinship T Kinship 16 16.0 1.000000 0.000000
T Soulmates T Soulmates 11 11.0 1.000000 0.000000
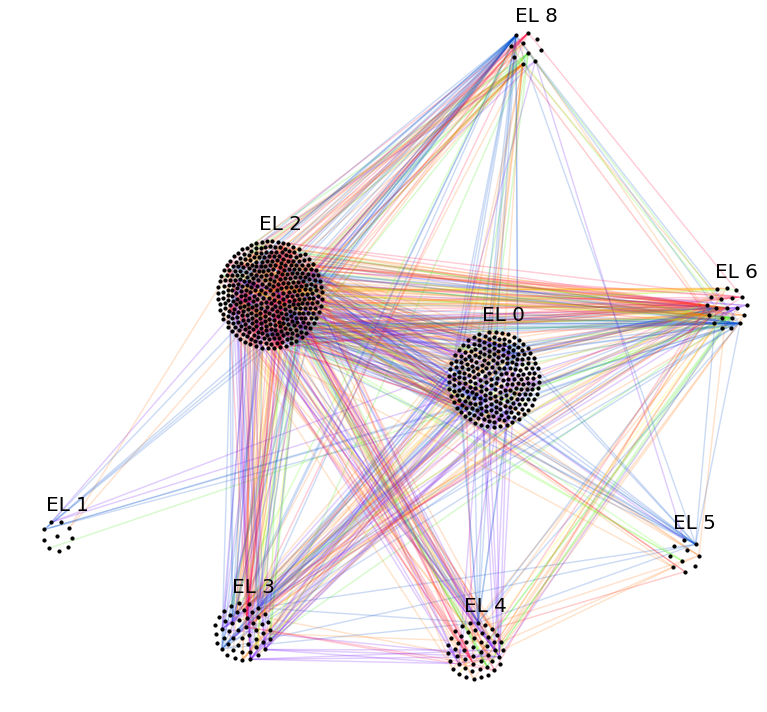
Let’s plot the graph g grouped by its layers.
# append layer_id to group nodes by layers
g.v['layer_id'] = g.v.groupby('layer').grouper.group_info[0].astype(np.int32)
# create graph_tool graph object
gtg = g.return_gt_graph(features=['layer_id'])
gtg.set_directed(False)
# get sfdp layout postitions
pos = gtd.sfdp_layout(gtg, groups=gtg.vp['layer_id'], mu=.15)
pos = pos.get_2d_array([0, 1])
g.v['x'] = pos[0]
g.v['y'] = pos[1]
# configure nodes
kwds_scatter = {'s': 10,
'c': 'k'}
# configure edges
kwds_quiver = {'headwidth': 1,
'alpha': .4,
'cmap': 'viridis'}
# color by weight
C = g.e.weight.values
# plot
fig, ax = plt.subplots(figsize=(12, 12))
obj = g.plot_2d('x', 'y', edges=True, C=C,
kwds_scatter=kwds_scatter,
kwds_quiver=kwds_quiver, ax=ax)
# turn axis off, set x/y-lim and name layers
ax.axis('off')
margin = 10
ax.set_xlim((g.v.x.min() - margin, g.v.x.max() + margin))
ax.set_ylim((g.v.y.min() - margin, g.v.y.max() + margin))
for layer in layer_names:
plt.text(g.v[g.v['layer'] == layer].x.mean() - margin * 3,
g.v[g.v['layer'] == layer].y.max() + margin,
layer, fontsize=15)
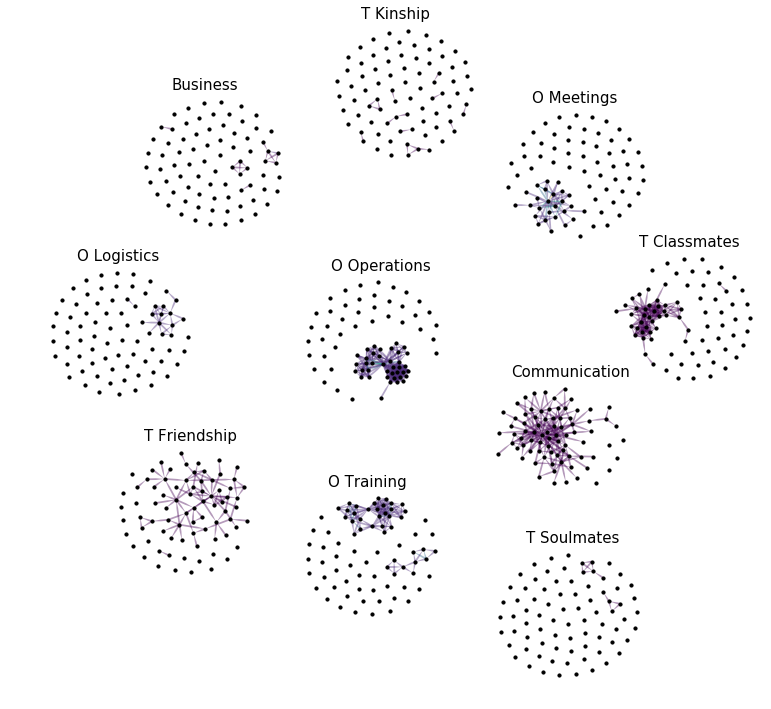
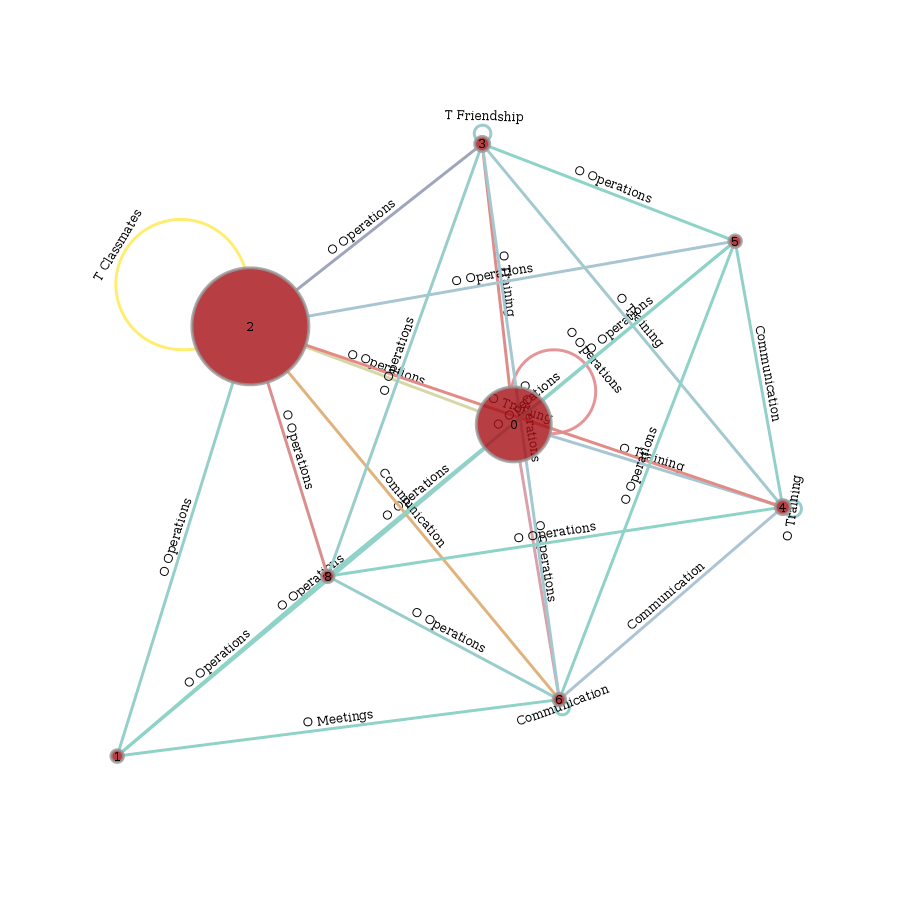
We can also plot the supergraph \(G^L = (V^L, E^L)\)
# create graph_tool graph of lg
gtg = lg.return_gt_graph(relations=True, node_indices=True, edge_indices=True)
# create plot
gtd.graph_draw(gtg,
vertex_text=gtg.vp['i'], vertex_text_position=-2,
vertex_fill_color='w',
vertex_text_color='k',
edge_text=gtg.ep['n_edges'],
inline=True, fit_view=.8,
output_size=(400,400))
The SuperGraph \(G^N = (V^N, E^N)\)¶
Partitioning by the type of feature V_N leads to the supergraph
\(G^{N} = (V^{N}, E^{N})\), where each supernode
\(V^{N}_{i^N} \in V^{N}\) corresponds to a node of the MLN.
Superedges \(E^{N}_{i^N j^N} \in E^{N}\) with \(i^N = j^N\)
correspond to the coupling edges of a MLN.
# partition by MLN's node indices
nv, ne, gv, ge = g.partition_graph('V_N', return_gve=True)
# for each superedge, get types of edges and their weights
def type_weights(group):
index = group['type'].values
data = group['weight'].values
return pd.Series(data=data, index=index)
ne_weights = ge.apply(type_weights).unstack()
ne = pd.concat((ne, ne_weights), axis=1)
# create graph
ng = dg.DeepGraph(nv, ne)
ng
<DeepGraph object, with n=79 node(s) and m=623 edge(s) at 0x7fb8d1da8b70>
print(ng.v)
n_nodes
V_N
0 10
1 10
2 10
3 10
4 10
.. ...
74 10
75 10
76 10
77 10
78 10
[79 rows x 1 columns]
print(ng.e)
n_edges Business Communication O Logistics O Meetings O Operations O Training T Classmates T Friendship T Kinship T Soulmates
V_N_s V_N_t
0 15 3 NaN 1.0 2.0 NaN NaN NaN NaN NaN 1.0 NaN
1 4 1 NaN NaN NaN NaN NaN NaN 1.0 NaN NaN NaN
5 1 NaN NaN NaN NaN NaN NaN 1.0 NaN NaN NaN
16 1 NaN NaN NaN NaN 2.0 NaN NaN NaN NaN NaN
21 1 NaN NaN NaN NaN NaN NaN 1.0 NaN NaN NaN
... ... ... ... ... ... ... ... ... ... ... ...
72 73 4 NaN 1.0 NaN NaN 2.0 2.0 NaN NaN 1.0 NaN
76 6 NaN 1.0 NaN 2.0 2.0 2.0 1.0 1.0 NaN NaN
77 2 NaN NaN 2.0 NaN NaN NaN NaN NaN NaN 1.0
73 76 2 NaN NaN NaN NaN 2.0 2.0 NaN NaN NaN NaN
75 78 2 NaN NaN NaN NaN NaN 2.0 NaN 1.0 NaN NaN
[623 rows x 11 columns]
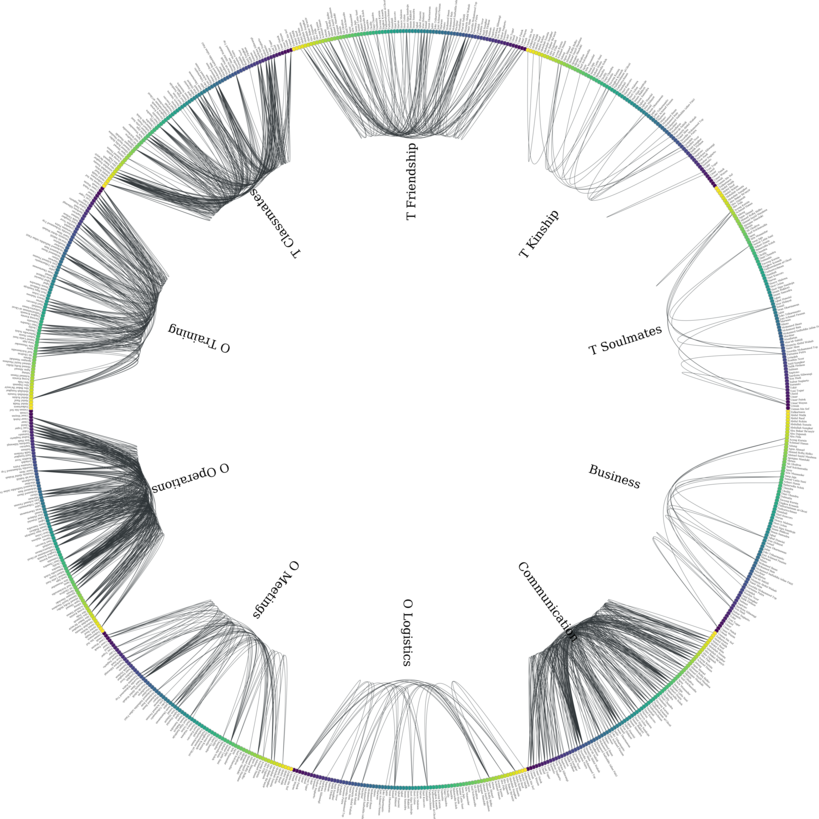
Let’s plot the graph g grouped by V_N.
# create graph_tool graph object
g.v['V_N'] = g.v['V_N'].astype(np.int32) # sfpd only takes int32
g_tmp = dg.DeepGraph(v)
gtg = g_tmp.return_gt_graph(features='V_N')
gtg.set_directed(False)
# get sfdp layout postitions
pos = gtd.sfdp_layout(gtg, groups=gtg.vp['V_N'], mu=.3, gamma=.01)
pos = pos.get_2d_array([0, 1])
g.v['x'] = pos[0]
g.v['y'] = pos[1]
# configure nodes
kwds_scatter = {'c': 'k'}
# configure edges
kwds_quiver = {'headwidth': 1,
'alpha': .2,
'cmap': 'viridis_r'}
# color by type
C = g.e.groupby('type').grouper.group_info[0]
# plot
fig, ax = plt.subplots(figsize=(15,15))
g.plot_2d('x', 'y', edges=True,
kwds_scatter=kwds_scatter, C=C,
kwds_quiver=kwds_quiver, ax=ax)
# turn axis off, set x/y-lim and name nodes
name_dic = {i: name for i, name in enumerate(vinfo.iloc[:79].Name)}
ax.axis('off')
ax.set_xlim((g.v.x.min() - 1, g.v.x.max() + 1))
ax.set_ylim((g.v.y.min() - 1, g.v.y.max() + 1))
for node in g.v['V_N'].unique():
plt.text(g.v[g.v['V_N'] == node].x.mean() - 1,
g.v[g.v['V_N'] == node].y.max() + 1,
name_dic[node], fontsize=12)
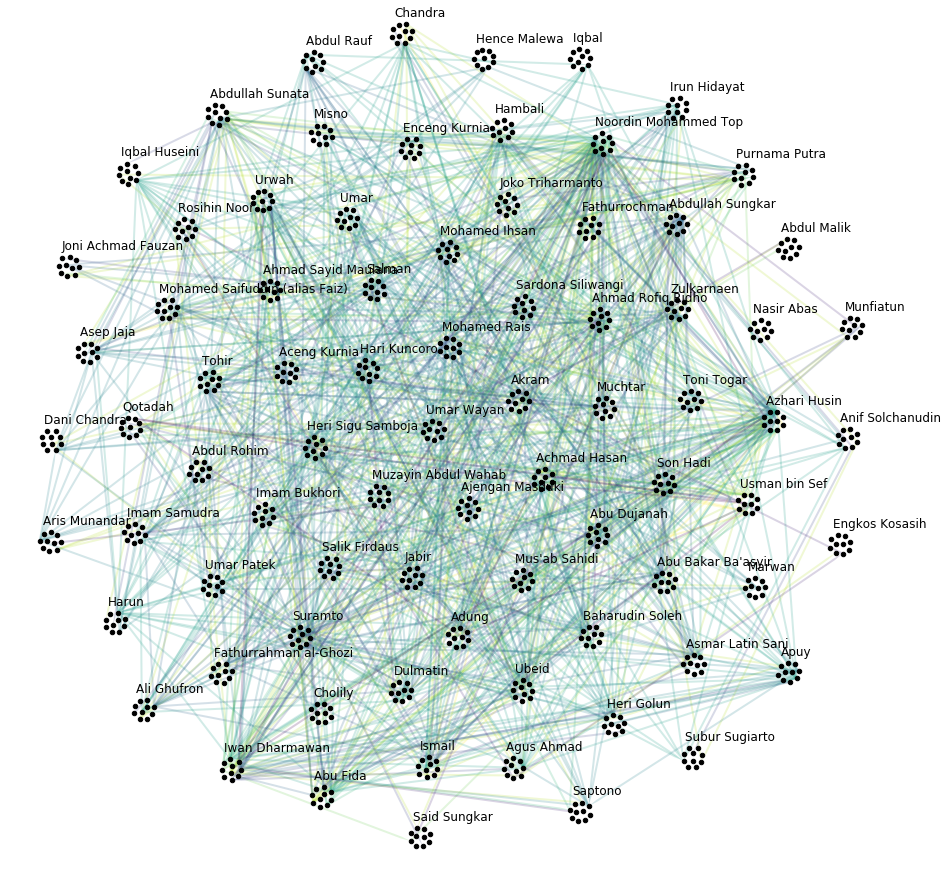
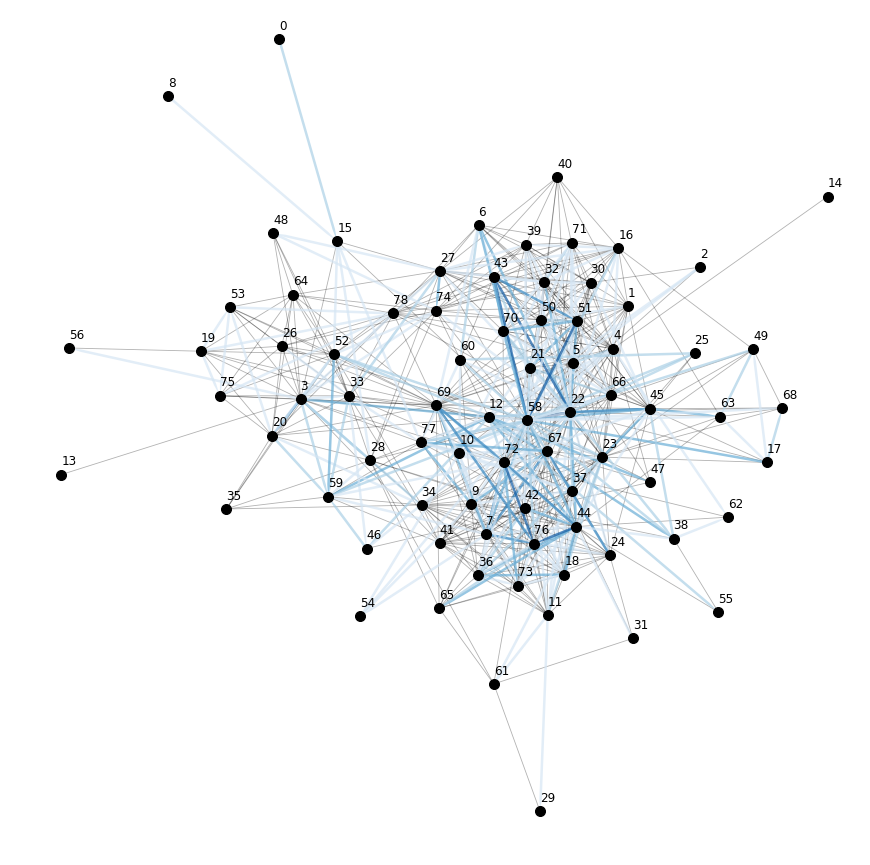
Let’s also plot the supergraph \(G^N = (V^N, E^N)\), where the color of the superedges corresponds to the number of edges within the respective superedge.
# get rid of isolated node for nicer layout
ng.v.drop(57, inplace=True, errors='ignore')
# create graph_tool graph object
gtg = ng.return_gt_graph(features=True, relations='n_edges')
gtg.set_directed(False)
# get sfdp layout postitions
pos = gtd.sfdp_layout(gtg)
pos = pos.get_2d_array([0, 1])
ng.v['x'] = pos[0]
ng.v['y'] = pos[1]
# configure nodes
kwds_scatter = {'s': 100,
'c': 'k'}
# configure edges
# split edges with only one type of connection
C_split_0 = ng.e['n_edges'].values.copy()
C_split_0[C_split_0 == 1] = 0
# edges with one type of connection
kwds_quiver_0 = {'alpha': .3,
'width': .001}
# edges with more than one type
kwds_quiver = {'headwidth': 1,
'width': .003,
'alpha': .7,
'cmap': 'Blues',
'clim': (1, ng.e.n_edges.max())}
# create plot
fig, ax = plt.subplots(figsize=(15,15))
ng.plot_2d('x', 'y', edges=True, C_split_0=C_split_0,
kwds_scatter=kwds_scatter, kwds_quiver_0=kwds_quiver_0,
kwds_quiver=kwds_quiver, ax=ax)
# turn axis off, set x/y-lim and name nodes
ax.axis('off')
ax.set_xlim(ng.v.x.min() - 1, ng.v.x.max() + 1)
ax.set_ylim(ng.v.y.min() - 1, ng.v.y.max() + 1)
for i in ng.v.index:
plt.text(ng.v.at[i, 'x'], ng.v.at[i, 'y'] + .3, i, fontsize=12)
The Tensor-Like Representation \(G^{NL} = (V^{NL}, E^{NL})\)¶
Considering only the information attributed to the layers of the MLN, and the fact that this MLN has just one aspect, there is only one more supergraph we can create of g. It is given by creating the intersection partition (see section III E of the Deep Graphs paper) of the types of features V_N and layer. The resulting supergraph \(G^{N \cdot L} = (V^{N \cdot L},E^{N \cdot L})\) corresponds one to one to the graph \(G = (V, E)\), and therefore to the supra-graph representation of the MLN. The only difference is the indexing, which is tensor-like for the supergraph \(G^{N \cdot L}\).
# partition the graph
relation_funcs = {'type': 'sum', 'weight': 'sum'} # just to transfer relations
nlv, nle = g.partition_graph(['V_N', 'layer'], relation_funcs=relation_funcs)
nlg = dg.DeepGraph(nlv, nle)
nlg
<DeepGraph object, with n=790 node(s) and m=1014 edge(s) at 0x7fb8d5325550>
print(nlg.v)
n_nodes
V_N layer
0 Business 1
Communication 1
O Logistics 1
O Meetings 1
O Operations 1
... ...
78 O Training 1
T Classmates 1
T Friendship 1
T Kinship 1
T Soulmates 1
[790 rows x 1 columns]
print(nlg.e)
n_edges weight type
V_N_s layer_s V_N_t layer_t
0 Communication 15 Communication 1 1.0 Communication
O Logistics 15 O Logistics 1 2.0 O Logistics
T Kinship 15 T Kinship 1 1.0 T Kinship
1 O Operations 16 O Operations 1 2.0 O Operations
22 O Operations 1 2.0 O Operations
... ... ... ...
72 T Soulmates 77 T Soulmates 1 1.0 T Soulmates
73 O Operations 76 O Operations 1 2.0 O Operations
O Training 76 O Training 1 2.0 O Training
75 O Training 78 O Training 1 2.0 O Training
T Friendship 78 T Friendship 1 1.0 T Friendship
[1014 rows x 3 columns]
This tensor-like index allows you to use the advanced indexing features of pandas.
print(nlg.e.loc[2, 'Communication', :, 'Communication'])
n_edges weight type
V_N_s layer_s V_N_t layer_t
2 Communication 5 Communication 1 1.0 Communication
12 Communication 1 1.0 Communication
30 Communication 1 1.0 Communication
58 Communication 1 1.0 Communication
In the future, we might implement a method to convert this tensor-representation of a MLN to some sparse-tensor data structure (e.g., https://github.com/mnick/scikit-tensor). Another idea is to create an interface to a suitable multilayer network package that implements the measures and models developed particularly for MLNs.
Partitioning Edges Based on Node Properties¶
Here, we demonstrate very briefly how to use the additional information of the nodes to perform queries on the edges.
# create "undirected" edge table (swap-copy all edges)
g.e = pd.concat((e, e.swaplevel(0,1)))
g.e.sort_index(inplace=True)
print(g.partition_edges(source_features=['Nationality']))
n_edges
Nationality_s
3 1655
4 351
5 22
print(g.partition_edges(source_features=['Nationality'], target_features=['Military Training']))
n_edges
Nationality_s Military Training_t
3 0 185
1 51
3 847
4 60
5 115
... ...
5 4 3
5 1
7 1
9 1
10 1
[26 rows x 1 columns]
print(g.partition_edges(source_features=['Nationality'],
target_features=['Military Training'],
relations='type'))
n_edges
type Nationality_s Military Training_t
Business 3 0 3
3 16
4 1
9 2
10 2
... ...
T Soulmates 3 9 1
10 2
4 3 3
9 1
10 3
[138 rows x 1 columns]
Alternative Representation of the MLN Edges¶
The edges of the supra-graph representation as presented in the paper look like this
print(e_paper)
Business Communication O Logistics O Meetings O Operations O Training T Classmates T Friendship T Kinship T Soulmates
s t
9 67 2.0 NaN NaN NaN NaN NaN NaN NaN NaN NaN
69 1.0 NaN NaN NaN NaN NaN NaN NaN NaN NaN
77 1.0 NaN NaN NaN NaN NaN NaN NaN NaN NaN
11 61 1.0 NaN NaN NaN NaN NaN NaN NaN NaN NaN
20 59 1.0 NaN NaN NaN NaN NaN NaN NaN NaN NaN
... ... ... ... ... ... ... ... ... ... ...
733 769 NaN NaN NaN NaN NaN NaN NaN NaN NaN 1.0
755 769 NaN NaN NaN NaN NaN NaN NaN NaN NaN 1.0
787 NaN NaN NaN NaN NaN NaN NaN NaN NaN 1.0
771 788 NaN NaN NaN NaN NaN NaN NaN NaN NaN 1.0
783 788 NaN NaN NaN NaN NaN NaN NaN NaN NaN 1.0
[1014 rows x 10 columns]
As you can see, the edge table is also comprised of 1014 edges between
the nodes in v. However, every type of connection get’s its own
column, where a “nan” value means that an edge does not have a relation
of the corresponding type.