10 Minutes to DeepGraph¶
[ipython notebook] [python script] [data]
This is a short introduction to DeepGraph. In the following, we demonstrate DeepGraph’s core functionalities by a toy data-set, “flying balls”.
First of all, we need to import some packages
# for plots
import matplotlib.pyplot as plt
# the usual
import numpy as np
import pandas as pd
import deepgraph as dg
# notebook display
%matplotlib inline
plt.rcParams['figure.figsize'] = 8, 6
pd.options.display.max_rows = 10
pd.set_option('expand_frame_repr', False)
Loading Toy Data
Then, we need data in the form of a pandas DataFrame, representing the nodes of our graph
v = pd.read_csv('flying_balls.csv', index_col=0)
print(v)
time x y ball_id
0 0 1692.000000 0.000000 0
1 0 8681.000000 0.000000 1
2 0 490.000000 0.000000 2
3 0 7439.000000 0.000000 3
4 0 4998.000000 0.000000 4
... ... ... ... ...
1163 45 2812.552734 16.503178 39
1164 46 5686.915998 14.161693 10
1165 46 3161.729086 19.381823 14
1166 46 5594.233413 57.701712 37
1167 47 5572.216748 20.588750 37
[1168 rows x 4 columns]
The data consists of 1168 space-time measurements of 50 different toy
balls in two-dimensional space. Each space-time measurement (i.e. row of
v) represents a node.
Let’s plot the data such that each ball has it’s own color
plt.scatter(v.x, v.y, s=v.time, c=v.ball_id)
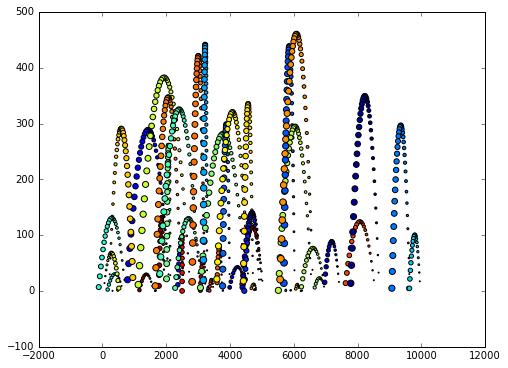
Creating Edges¶
In order to create edges between these nodes, we now initiate a dg.DeepGraph instance
g = dg.DeepGraph(v)
g
<DeepGraph object, with n=1168 node(s) and m=0 edge(s) at 0x7facf3b35dd8>
and use it to create edges between the nodes given by g.v. For that matter, we may define a connector function
def x_dist(x_s, x_t):
dx = x_t - x_s
return dx
and pass it to g.create_edges in order to compute the distance in the x-coordinate of each pair of nodes
g.create_edges(connectors=x_dist)
g
<DeepGraph object, with n=1168 node(s) and m=681528 edge(s) at 0x7facf3b35dd8>
print(g.e)
dx
s t
0 1 6989.000000
2 -1202.000000
3 5747.000000
4 3306.000000
5 2812.000000
... ...
1164 1166 -92.682585
1167 -114.699250
1165 1166 2432.504327
1167 2410.487662
1166 1167 -22.016665
[681528 rows x 1 columns]
Let’s say we’re only interested in creating edges between nodes with a x-distance smaller than 1000. Then we may additionally define a selector
def x_dist_selector(dx, sources, targets):
dxa = np.abs(dx)
sources = sources[dxa <= 1000]
targets = targets[dxa <= 1000]
return sources, targets
and pass both the connector and selector to g.create_edges
g.create_edges(connectors=x_dist, selectors=x_dist_selector)
g
<DeepGraph object, with n=1168 node(s) and m=156938 edge(s) at 0x7facf3b35dd8>
print(g.e)
dx
s t
0 6 416.000000
7 848.000000
19 -973.000000
24 437.000000
38 778.000000
... ...
1162 1167 -44.033330
1163 1165 349.176351
1164 1166 -92.682585
1167 -114.699250
1166 1167 -22.016665
[156938 rows x 1 columns]
There is, however, a much more efficient way of creating edges that involve a simple distance threshold such as the one above
Creating Edges on a FastTrack¶
In order to efficiently create edges including a selection of edges via a simple distance threshold as above, one should use the create_edges_ft method. It relies on a sorted DataFrame, so we need to sort g.v first
g.v.sort_values('x', inplace=True)
g.create_edges_ft(ft_feature=('x', 1000))
g
<DeepGraph object, with n=1168 node(s) and m=156938 edge(s) at 0x7facf3b35dd8>
Let’s compare the efficiency
%timeit -n3 -r3 g.create_edges(connectors=x_dist, selectors=x_dist_selector)
3 loops, best of 3: 557 ms per loop
%timeit -n3 -r3 g.create_edges_ft(ft_feature=('x', 1000))
3 loops, best of 3: 167 ms per loop
The create_edges_ft method also accepts connectors and selectors as input. Let’s connect only those measurements that are close in space and time
def y_dist(y_s, y_t):
dy = y_t - y_s
return dy
def time_dist(time_t, time_s):
dt = time_t - time_s
return dt
def y_dist_selector(dy, sources, targets):
dya = np.abs(dy)
sources = sources[dya <= 100]
targets = targets[dya <= 100]
return sources, targets
def time_dist_selector(dt, sources, targets):
dta = np.abs(dt)
sources = sources[dta <= 1]
targets = targets[dta <= 1]
return sources, targets
g.create_edges_ft(ft_feature=('x', 100),
connectors=[y_dist, time_dist],
selectors=[y_dist_selector, time_dist_selector])
g
<DeepGraph object, with n=1168 node(s) and m=1899 edge(s) at 0x7facf3b35dd8>
print(g.e)
dt dy ft_r
s t
890 867 -1 19.311136 33.415831
867 843 -1 17.678482 33.415831
843 818 -1 16.045829 33.415831
818 792 -1 14.413176 33.415831
792 766 -1 12.780523 33.415831
... .. ... ...
244 203 -1 -10.825226 15.455612
203 159 -1 -12.457879 15.455612
159 114 -1 -14.090532 15.455612
114 65 -1 -15.723185 15.455612
65 16 -1 -17.355838 15.455612
[1899 rows x 3 columns]
We can now plot the flying balls and the edges we just created with the plot_2d method
obj = g.plot_2d('x', 'y', edges=True,
kwds_scatter={'c': g.v.ball_id, 's': g.v.time})
obj['ax'].set_xlim(1000,3000)
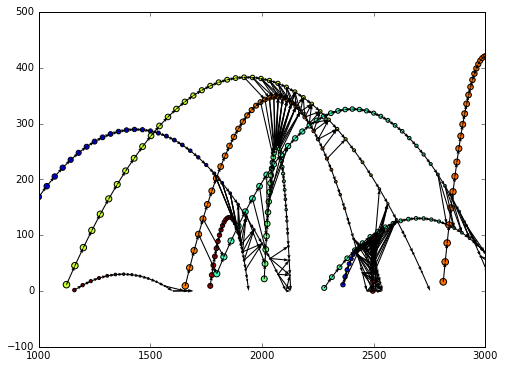
Graph Partitioning¶
The DeepGraph class also offers methods to partition nodes, edges and an entire graph. See the docstrings and the other tutorials for details and examples.
Graph Interfaces¶
Furthermore, you may inspect the docstrings of return_cs_graph, return_nx_graph and return_gt_graph to see how to convert from DeepGraph’s DataFrame representation of a network to sparse adjacency matrices, NetworkX’s network representation and graph_tool’s network representation.
Plotting Methods¶
DeepGraph also offers a number of useful Plotting methods. See plotting methods for details and have a look at the other tutorials for examples.